Can a 'Google AI' Build Your Genome Sequence?
A new AI-powered tool from Google promises more-accurate genome sequences, but its impact on genomics research remains to be seen
What Happened
In December 2017, Google announced a new tool called DeepVariant:
Today, we announce the open source release of DeepVariant, a deep learning technology to reconstruct the true genome sequence from HTS sequencer data with significantly greater accuracy than previous classical methods.
Simultaneously, the DeepVariant authors published a paper detailing their research and, unlike many research projects, provided the complete underlying source code — yay! So what is it, and how does it work?
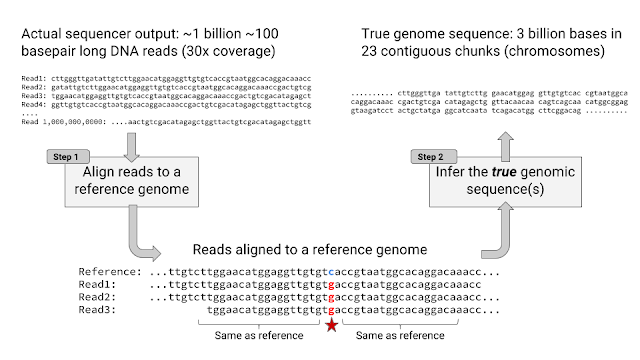
First, let’s talk about DNA. DNA is the molecule that determines your genetic makeup, influencing everything from your height to your eye color to your risk of getting cancer. Each person’s genetic information can be represented as a sequence of the letters A, T, C, and G. Thanks to a technology called high-throughput sequencing, it is now possible to read the ~3 billion letters in your DNA for just ~$1000, which has in turn been fueling the rapidly growing field of genomics research.
Here’s how high-throughput sequencing typically works: DNA molecules are isolated from blood or spit and broken into millions of pieces with a couple hundred letters each. To read each letter, a machine adds a chemical dye to each piece, lights up the dye with a laser, and takes a picture. The rest of the process happens on computers — a program processes each picture to determine the most likely letter sequence for each piece, aligns each piece to a complete reference sequence, and lists “variants” (a variant is any difference between the sample and reference sequence e.g. “three letters deleted” or “A instead of T”).
DeepVariant performs only the last step — determining which variants the aligned pieces represent. It’s powered by Deep Learning, the technique behind most recent advances in AI and machine learning. As the authors demonstrate, it is significantly more accurate than existing tools, making 10x fewer errors. Interestingly, DeepVariant is based on a deep neural network architecture that was initially developed to classify whether a picture contains objects like dogs or cats, showcasing how the same Deep Learning approach can solve wildly different problems.
The Reactions
The media coverage about DeepVariant was generally accurate in its portrayal of both the technical and non-technical aspects of the research:
-
Wired reported that “Google Is Giving Away AI That Can Build Your Genome Sequence”. The article does a great job of providing context for the research and the problem it solves, as well as telling the human story about how the project started at Google.
-
The MIT Technology Review published an article with the more-muted title “Google Has Released an AI Tool That Makes Sense of Your Genome”. It goes more into detail about the competitive landscape for these tools instead of focusing on the human story.
-
The Atlantic used the engaging headline “Google Taught an AI That Sorts Cat Photos to Analyze DNA”. This is technically correct, since DeepVariant uses the same neural network architecture that excels at other image processing problems.
-
A Forbes blog post titled “No, Google’s AI Program Can’t Build Your Genome Sequence” called out Wired for their use of the phrase “assembling genomes” (genome assembly is a different problem in genomics, and not the one DeepVariant solves). But this is really a nitpick — to a general audience, it’s a reasonable description of what DeepVariant does, although a technical audience may interpret the word “assembling” differently. Wired nonetheless clarified this in a correction.
Many headlines made the mistake of calling DeepVariant an “AI” (artificial intelligence), when it is really a computer program that uses AI techniques to determine genetic variants. It is not an “intelligence” in itself. Besides this common wording mistake, the coverage was on the whole well done.
Our Perspective
So a 10x reduction in errors is awesome, right? Yes, but with two catches. First, even though DeepVariant reduces the number of errors by 10x, there are relatively few errors to begin with — in one test, DeepVariant achieved an accuracy score of 0.99, while its competitors are not far behind at ~0.97. Second, DeepVariant is deeply expensive — it requires between 2 and 13 times as much computational power as its competitors. Doubling your computational budget is a difficult ask of research groups that operate on fixed grants, particularly for a marginal increase in accuracy.
A search of Google Scholar reveals that as of March 2018, no scientific publications have used it except for testing. Meanwhile, over 400 papers in 2018 used the competing framework GATK and over 700 used SAMtools. It appears that computational cost or general unfamiliarity are deterring its adoption in the research community. In fairness, DeepVariant has only been available for three months, and adoption may pick up over time as specialized AI processors become more available to academic researchers. It’s also possible the tool could be made faster without losing accuracy or see greater adoption through Google cloud offering of it.
In some ways, DeepVariant perfectly illustrates the strengths and weaknesses of Deep Learning. Unlike previous tools, it doesn’t require a team of experts to spend years teaching it to deal with different kinds of errors — it simply learns patterns by being shown lots of pictures. However, it requires much more computer power and only incrementally improves accuracy.
TLDR
Google’s DeepVariant is a more-accurate method for doing one part of the genome sequencing process. It has seen little adoption by researchers in its first three months, likely due to the extra computational cost for marginal improved accuracy. Media coverage correctly describes the research, but its impact on the broader scientific field remains to be seen.
Disclosure: I worked for Google and still have some equity.







